Editor’s note: Several small technical corrections have been made to this article since it was first published.
GENOMIC testing has opened a range of new opportunities for seedstock and commercial producers. For a number of larger breeds, genomics has been incorporated into the data used to generate Breeding Values, for breeds enrolled in either Breedplan or IGS. The addition of genomic information has resulted in significant improvements in accuracy of data which has not just enhanced sire selection but has been used to select replacement females as well.
 The release of Genomic Breeding Values (reported previously in Beef Central) by QAAFI has taken the application of genomics a further step into commercial use, offering producers the opportunity to benchmark herd performance on a range of fertility traits.
The release of Genomic Breeding Values (reported previously in Beef Central) by QAAFI has taken the application of genomics a further step into commercial use, offering producers the opportunity to benchmark herd performance on a range of fertility traits.
Similar products have been offered through consultancies such as Thomas Elder Consulting & Neogen for commercial crossbred cattle.
While genomics is an area where interest and adoption are growing, there are limitations. These are important to note, particularly for breeds that are currently not using or have low levels of performance recording and are considering these products as a method to explore in lieu of traditional genetic information such as EBVs.
Perhaps the greatest limitation is the requirement for an accurate and reliable reference population. A reference population is comprised of animals from across the breed that are measured and recorded for the economically valuable traits that are reported in programs such as Breedplan. These records, along with the information recorded as part of the normal run of data used to develop EBVs (or EPDs in programs such as IGS), combine to enhance the accuracy and reliability of the performance data offered on animals.
It is essential to note that a reference population must have data on both the animals genotype and its phenotype. According to Dr Matt Wolcott of AGBU Without the recording of phenotype, “it really doesn’t matter how many genotypes you have or how heritable the trait is. Genomic breeding values are simply a prediction of the genetic component of performance from SNO information (lots of 0s, 1s and 2s). If you don’t have the phenotypes, you have no basis for predicting the relationship between all those 0s, 1s and 2s and genetic performance.”
It is also extremely important when considering this approach to recognise that these accuracy estimates only apply when the reference population is from the same population and is related to the animals for which the genomic breeding values are being estimated.
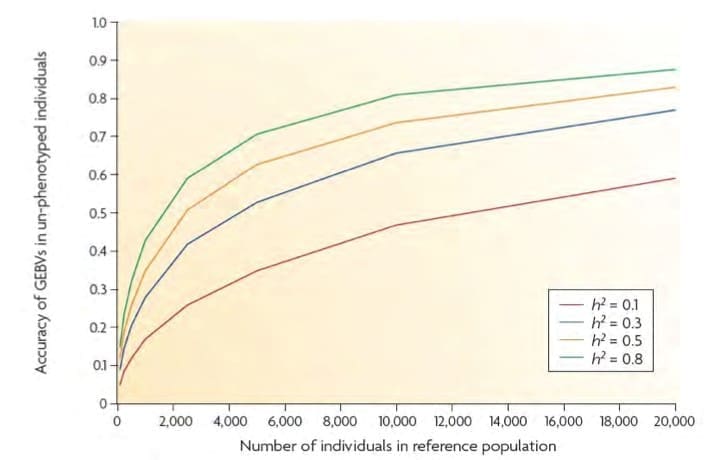
Source: Goddard & Hayes 2008
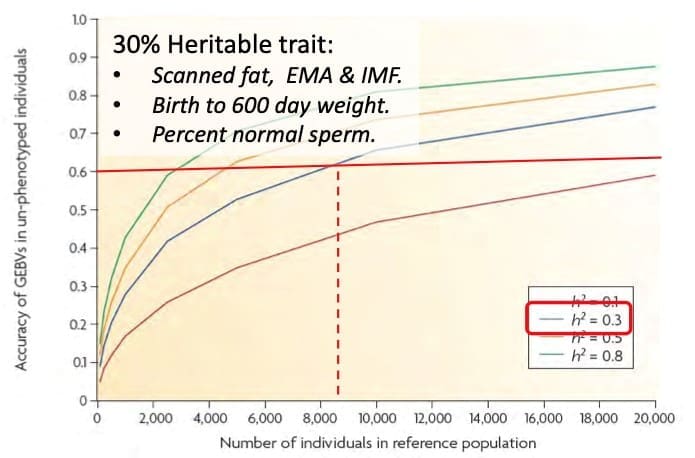
Figure 1 presents an estimate of EBV accuracy (on the vertical axis) which can be expected for animals with genotypes but no phenotype, when genomic breeding values are estimated based on related reference populations of 0-20K animals (on the horizontal axis). The green, yellow, blue and red curves are for traits with heritabilities of 0.1, 0.3, 0.5 and 0.8, and demonstrate the need for larger reference populations as trait heritabilities decline, with the size of the reference required to achieve 60% accuracy in genotyped only animals for a 0.3 heritable trait illustrated by the red lines (just over 8,ooo reference animals).
It is important to realise that these estimates can only be reliable when there is a relationship between the reference population and the animals which have had only genomic information collected for analysis, and the closer those relationships the greater the accuracies will become.
Without a relationship between the reference population and the target animals, expectations for EBV accuracy must be extremely low, regardless of how many genotypes are included in the analysis.
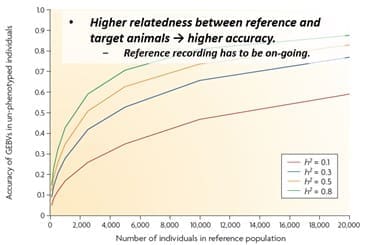
For smaller breeds achieving a suitable reference population of related animals may actually be well beyond the physical size of the bred as they currently stand. In addition the reference population needs to have on going recording as part of the data used in the development of any EBVs.
As is the case for all effective genetic evaluations, its essential that the data collected is actually based on economically important traits and that there is an ongoing commitment to recording and maintaining the data. In simple terms genetic testing without an accurate and relevant reference population is unlikely to be of great usefulness to a breed. It certainly won’t offer a short cut to highly accurate EBVs.
Breeders and producers considering the role genomics may play in their herd or breed should perhaps consider if there is firstly sufficient data being collected to be used to generate meaningful breeding values. If the answer is no, a more valuable process may be to bring recording of phenotypes to a standard where it can be used to generate that information. From there, the inclusion of genotypes to enhance accuracies and to offer on farm decisions may actually become a viable option.
 Alastair Rayner is the Principal of RaynerAg, an agricultural advisory service based in NSW. RaynerAg is affiliated with BJA Stock & Station Agents. He regularly lists and sell cattle for clients as well attending bull sales to support client purchases. Alastair provides pre-sale selections and classifications for seedstock producers in NSW, Qld and Victoria. He can be contacted here or through his website www.raynerag.com.au
Alastair Rayner is the Principal of RaynerAg, an agricultural advisory service based in NSW. RaynerAg is affiliated with BJA Stock & Station Agents. He regularly lists and sell cattle for clients as well attending bull sales to support client purchases. Alastair provides pre-sale selections and classifications for seedstock producers in NSW, Qld and Victoria. He can be contacted here or through his website www.raynerag.com.au



Hi Alastair, thanks for an interesting article. When assessing which genomic tools to use, we would also encourage people to ask the question “Is the reference population used to predict the genomic breeding values relevant to my production system?”. For example, a reference population using only US data may not be particularly relevant for some Australian production systems. The result would be genomic breeding values that may not be very accurate when used to select for performance in Australian conditions, depending on how different the production environments are. Reference data collected under Australian conditions would result in higher accuracies of genomic breeding values for performance in Australian conditions.