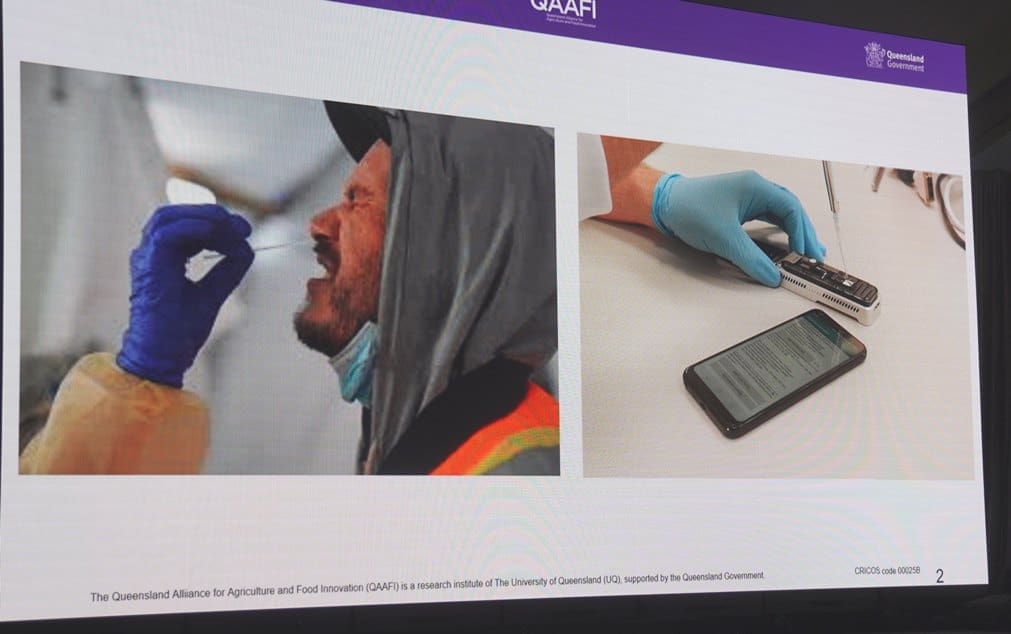
New portable DNA sequencing systems can deliver a COVID test result in 15 minutes, and have considerable potential in cattle breeding and selection
BREAKTHROUGHS in diagnostic technology targeted at human health during the COVID era are paving the way for ever-more rapid progress in selection of beef cattle for important productivity and quality traits.
During an MLA genetics seminar held during Beef 2021, QAAFI geneticist Prof Ben Hayes provided an exciting glimpse of the future in terms of tools likely to become available to assist beef producers in selecting for more fertile, efficient, better-eating cattle.
“As anybody who has had a COVID test may know, the time between the test being taken and the result being returned has reduced quite remarkably over the past year,” Prof Hayes said.
“When I first had a COVID test, I had to wait two days in isolation until I got the result back. The last test I had, it was 45 minutes. Clinics in Sydney now can make a diagnosis in 15 minutes.”
Some of that progress has been the result of the device pictured above – a portable hand-held genome sequencer.
“Place a drop of DNA in the device, and it can quickly sequence the entire genome of an organism,” Prof Hayes said, “and that organism can be a COVID 19 virus, a human being, or in our case, a cow.”
Researchers had also found that the test could be sped-up even further by focusing only a key ‘bits’ of the COVID virus genome.
“For cattle, that is going to be really important in the future, because the cattle genome is quite big – three billion ‘As, Cs, Ts and Gs’. If we can just focus on certain bits of the cattle genome, we can use this little device to sequence DNA markers and so on, and do it really quickly,” Prof Hayes said.
Accurately determining animal age in extensive herds
Prof Hayes focused on the challenges faced in the northern beef industry to illustrate where this, and other new technologies could be used.
“Many large northern cattle properties muster their cattle only once, or maybe twice a year. That is their only chance to get some DNA information,” he said.

Prof Ben Hayes
“In many of those herds where bulls are in the breeder paddock for long periods, owners have little idea how old each calf is. They could be anything from one to four (months) old – and a starting point for BreedPlan recording is a reasonably accurate reference to date of birth.
Another challenge in large, extensive breeding herds is determining fertility in – sorting out the repeat calvers from those likely to have only one or two calves during their productive life, costing the producer money.
“The aim is to identify those non-performers as early as possible, because the infertile breeders are not providing any return. If we could identify those with good fertility genetics early on, producers could make considerable savings. Put that on the bull side as well, and the fertility of their daughters is also improved,” Prof Hayes said.
Epigenetics
The term, epigenetics surfaced at a number of seminars during Beef 2021, and Prof Hayes raised it again in this forum, describing it as the switching on or off of genes, as a result of environmental cues.
“Participants at this breakfast seminar eating their breakfast roll, are sending a signal to their gut to start the digestion process. That’s an example of epi-genetics,” he said.
Prof Hayes said that as living organisms aged, there was a systematic switching on and off of different genes over time.
“As people age, the body thinks it does not need certain genes anymore, so they get switched off. It turns out that ‘switching-off’ is very methodical – month after month, year after year, certain genes get switched off, in a very methodical fashion.”
In humans, that trait has been used to develop what is called the epi-genetic, or methylation ‘clock’, which has proven to be quite accurate in determining age. The assessment is being used for all kinds of applications, including forensics at crime scenes, where a hair or blood sample can be used to accurately determine how old the perpetrator of the crime was.
“We’d like to use this concept in cattle, to more accurately determine how old our animals are – especially those run in large northern herds, where knowledge about age is much more limited,” Prof Hayes said.
It turns out the same technology that is being used so widely in the COVID epidemic, called a nonopore sequencer, can also record these methylation profiles.
So how accurate are they likely to be in determining age of cattle?
An experiment was set up using a reference cattle population, using animals of known age.
Using about 70 cattle from five northern herds, tail hairs were extracted, and put through the nanopore sequencer.
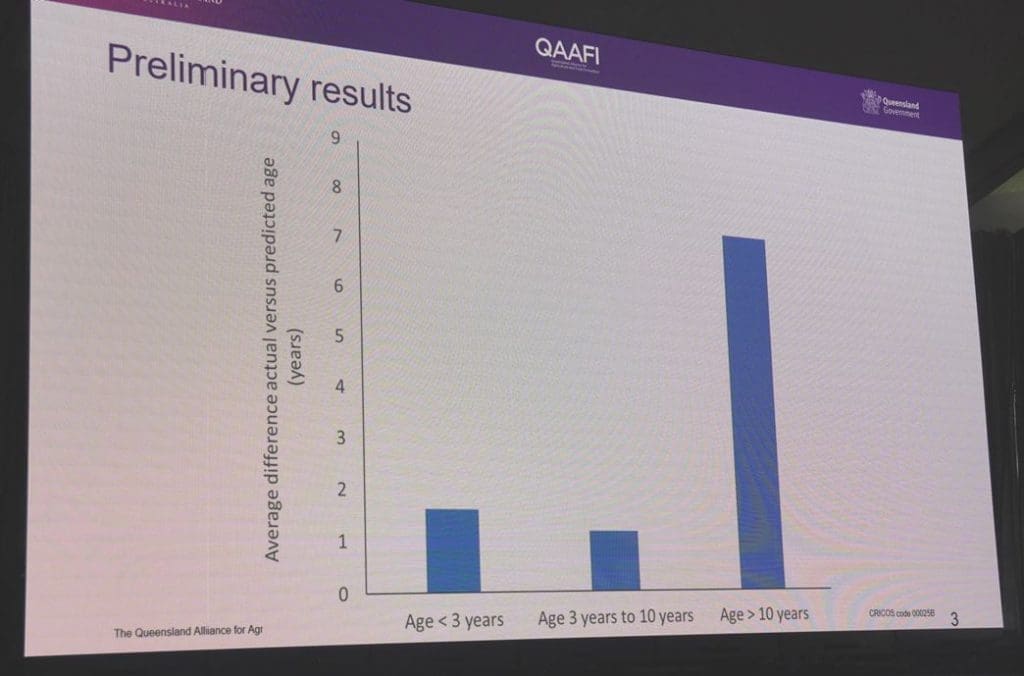
As the graph below shows, for animals older than ten years, the technology did a ‘really poor job’ of predicting age – although part of this was linked to the fact that was only a small sample of five animals involved. “We generally got that they were older than ten years, but within that, the resolution was pretty poor,” Prof Hayes said.
But for the class of animals that is more interesting, from a beef production perspective – animals between three and ten years – the accuracy improved dramatically.
“The first trial suggested we can already determine actual age within a year, in this age group,” Prof Hayes said. For the group less than three years of age, it was about the same.
Prof Hayes said there was still ‘some way to go’ with the age determination research, suggesting the final objective was to get accuracies within one month of actual birth date.
“Even within one year of birth-date is still useful for some purposes in northern cattle conditions, but for genetic evaluation purposes, we want get accuracy within one month of birth. As a result of doing this experiment, we now know how to get there – we just need more animals in our future experiments – probably a thousand or more, instead of less than a hundred.”
“But the good news is we can start to predict something about when an animal was born – and that blows me away, that we can do that from a tail-hair sample.”
In other applications, the age test could potentially play a role similar to ossification in carcase assessment, in predicting tenderness in eating quality in carcases.
Crush-side genotyping
The relentless focus on fertility in northern genetics was evident at numerous seminars held during Beef 2021, where all the dollar indexes for live export cattle or Jap Ox illustrate the big impact that fertility has on northern profitability.
Researchers were focusing heavily on whether genetic values could be predicted for fertility in northern cattle.
The first tool that’s needed to look at this was a reference population. The northern genomics project is providing that, with 30,000 heifers assessed out of 54 co-operator herds, followed through their productive life. All were DNA tested.
Traits examined include heifer puberty, which is closely correlated with lifetime calving, preg testing year after year, re-breed information for heifers and cows, body condition, height and other traits.
“But our ultimate aim is to enable DNA testing, and the production of breeding values, to be done on farm,” Prof Hayes said.
At the moment, to get this done, tail hairs or ear-notches are extracted and sent away to a lab, where they are genotyped (reading off the As, Cs Gs and Ts), which can take 6-8 weeks before the results come back.
“That’s no good if a producer is mustering his animals only once a year – he or she wants that information straight away.”
One of Prof Hayes’ PhD students Harrison Lamb, is working on the application of crush-side genotyping, using a portable DNA sequencer – exactly the same device that is currently being used to diagnose COVID in humans.
It is plugged into the laptop, the DNA extracted from the animal in the crush and put into the device, which gives back GBVs (genomic breeding values) within minutes, allowing producers to rank animals and make decisions on which to keep or cull.
“We certainly haven’t got there yet, but we’ve shown that the sequence coming out of this little device is just as accurate as the current technology – the SNP chip test genotyping done in the labs.
“It’s very encouraging – at least in the lab. So far this technology is working very well.”
Another researcher, Elizabeth Ross, has shown the crushside sequencer can produce parentage with 99.9pc accuracy.
“We hope in a couple of years after on-farm testing to be able to deliver a nice, streamlined system that can deliver genomic breeding values, parentage, age-profile – all from a crushside test.”




Absolutely brilliant forward movement for the industry. The inventors, developers and adaptors to the cattle industry of the portable DNA sequencer deserve a big round of applause.
Awesome potential. Hope they have the funds and people to speed up developments.